SEC-SAXS Instructions the basics
Introduction Registration Samples Experimental Details Shipping Data Resources CitingIntroduction mail-in SEC-SAXS at SIBYLS
SEC-SAXS workflow
Below is a schematic diagram of the Size-Exclusion Chromatography Small Angle X-ray Scattering (SEC-SAXS) workflow that you may take advantage of at the SIBYLS beamline 12.3.1 at the Advanced Light Source in Berkeley:
Coupling SEC with SAXS
Separation of analytes is typically accomplished by size-exclusion chromatography (SEC) using an Agilent 1260 series HPLC with a Shodex analytical column at a flow rate of 0.65 ml/min using your favorite buffer with a maximum of pH 7.5. The eluent from SEC is measured in line with 1) MALS and UV, 2) SAXS, 3) Differential refractivity.
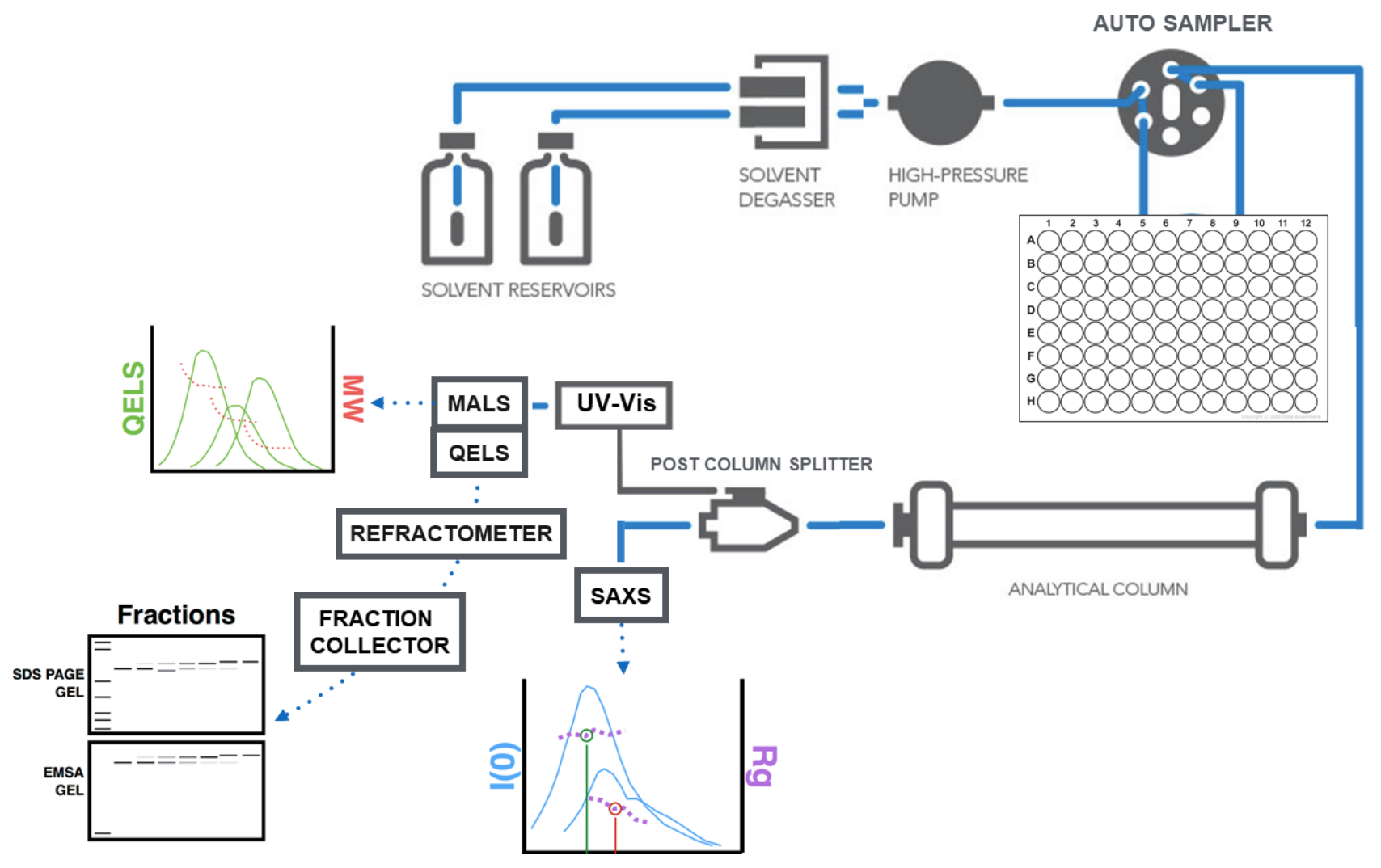
You can use this template for a detailed methods section in your publications.
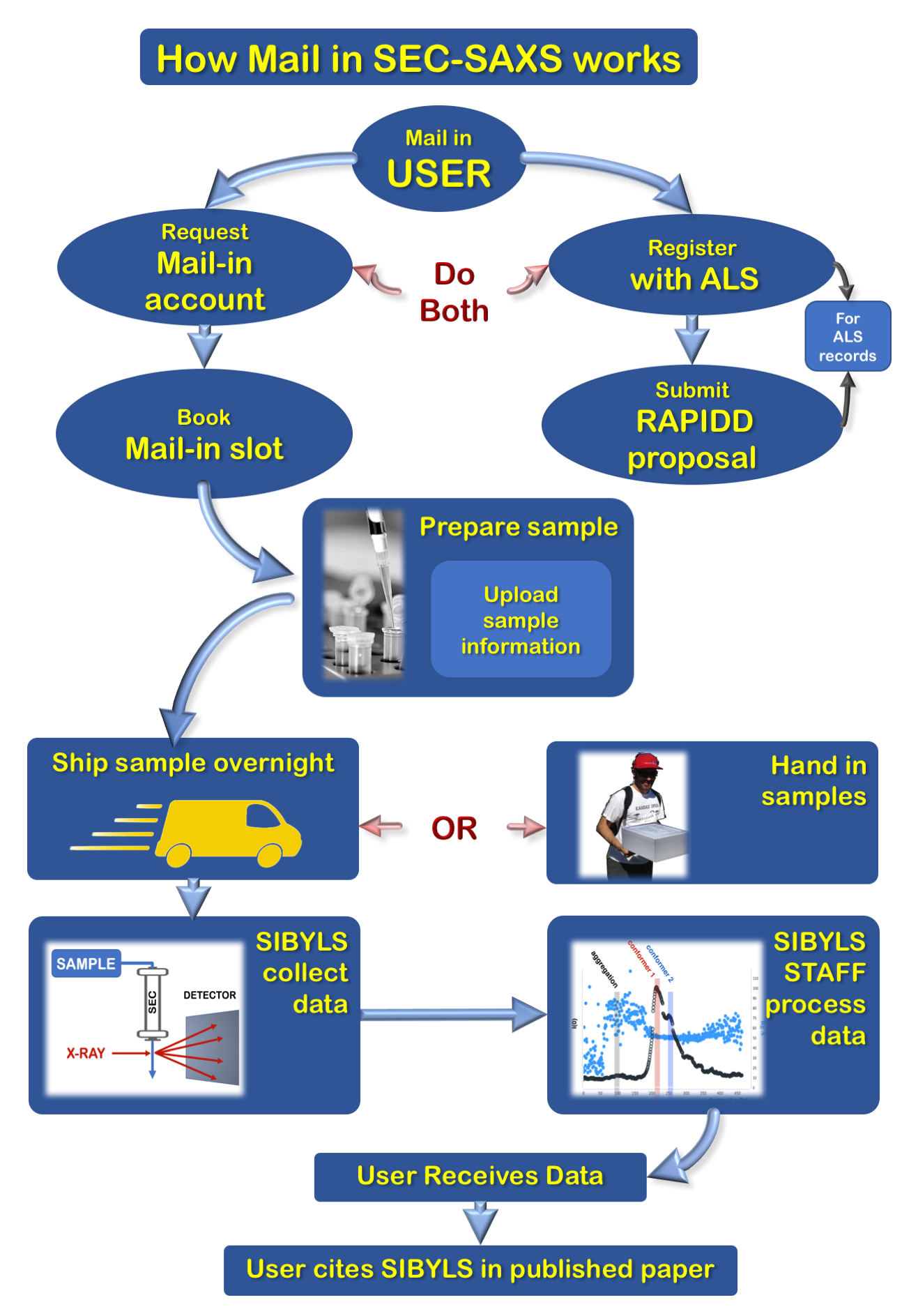
Registration request beamtime in 4 easy steps
1. Request mail-in account
All SIBYLS MailinSAXS users must first register for a MailinSAXS account
Request Mail-in Account2. Register with ALS & Submit a RAPIDD Proposal
All users must register with the ALS User Office, and submit one ALS RAPIDD proposal for every three MailinSAXS slots
3. Register at Simple Scattering
To expedite processing and sending your data, your data will be sent to Simple Scattering. Please register by clicking the button below
Simple Scattering4. Book mail-in slot
After completing the previous 3 steps, you may book a mail-in slot for beamtime
Book Mail-in SlotSamples information on sample, column and buffer requirements
Preparing your samples and buffer
Each beamtime slot is limited to 7 samples using 1 column and 1 buffer.
Each column run has the following sample and buffer requirements:
- A sample with a minimum volume of 60 µl
- 50 ml of 10x SEC buffer
The sample concentration should be from 3-10 mg/ml with smaller molecules requiring higher concentrations. Ideally, the sample should be prepared in a buffer that is identical or very similar to the SEC buffer.
For SEC buffers, the salt concentration must not exceed 500 mM and the pH of the buffer must be less than 7.5 to be accepted. We highly recommend adding 10-20% glycerol in your 10X buffer, so when diluted with dd H2O your 1x buffer will have 1-2% glycerol.
Please use small concentrations of other stabilizers such as glucose, ATP, etc. to minimize scattering background and to avoid high column-pressures.
Size-exclusion columns
A list of available columns with a summary of their properties are shown in the Table below. Please specify the columns to use in your experiments.
| Product Name | Exclusion Limit (Protein kDa) | Particle Size (μm) | Maximum Pore Size (Å) | Column Size (mm) ID x L |
|---|---|---|---|---|
| PROTEIN KW-802.5 | 150 | 5 | 400 | 8 x 300 |
| PROTEIN KW-803 | 700 | 5 | 400 | 8 x 300 |
| PROTEIN KW-804 | 1000 | 7 | 1500 | 8 x 300 |
Experimental Details buffer, column and sample entry
Entering experimental and sample information
Once you book an SEC slot, you will be prompted to enter the experimental details for your SEC-SAXS data collection. This includes entering the buffer you will supply, a choice of our available columns, and up to 7 samples.
You can find all of your slots by clicking the "My Account" tab that appears after logging in.
Shipping how to package and ship your samples and buffers
Ship sample overnight
We recommended that samples are shipped in standard Eppendorf PCR tubes together with 10x buffer using 4 °C cold packs (NOT frozen) in a styrofoam shipping container. Tightly pack the styrofoam box by surrounding assembly with more cold packs. While in some cases it is ok if the packs are frozen, do not allow the tubes to touch them.
DO NOT ship wet ice.
Clearly label the outside of your shipping box with the temperature of your samples. This is extremely important if a shipping delay occurs.
Lastly, please provide us with a shipping tracking number.
Shipping address
Ship samples overnight to arrive at SIBYLS on the first day of your shift:
- Lawrence Berkeley Lab
- ATTN: Kathryn Burnett / SIBYLS 12.3.1
- 1 Cyclotron Road MS 6R2136
- Berkeley, CA 94720
- Phone: (510) 495-3116
SHIPPING ADDRESS:
Data what happens after your sample arrives
Results and Analysis
We will provide a merged SAXS curve for each sample with a short report that includes SAXS parameters and a P(r) function to assess the quality of your data. We will also provide an SEC-MALS report for further quality control. Your results will be deposited to SIMPLE SCATTERING.
Please use the program RAW for viewing your merged SAXS profiles (.dat), derived P(r) functions (.out ), SAXS parameters, or SEC-SAXS chromatograms (.hdf5 ).
For more information please visit our tutorials. You can also choose the amount of SIBYLS staff involvement in processing and analyzing the results. Our team uses a variety of programs and decades of combined experience to help you with your analysis.
Feedback
We are continuing to improve our systems with the aim of making data of higher quality. Please provide us with constructive feedback. For additional information please feel free to contact:
- Michal Hammel, Ph.D
- SAXS Scientist
- Mhammel@lbl.gov
Citing the key to keep SIBYLS running to serve you
Cite SIBYLS in published papers
Please cite all relevant SIBYLS beamline papers below in the 'References' section of your journal article or book chapter
- Rosenberg DJ, Hura GL, Hammel M (2022) "Size exclusion chromatography coupled small angle X-ray scattering with tandem multiangle light scattering at the SIBYLS beamline" Meth. enzymol. 677, 191-219. (SEC-SAXS)
- Classen S, et al. (2013) “Implementation and performance of SIBYLS: a dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source” J. Appl. Crystallogr. 46(Pt 1), 1-13. (SIBYLS Beamline hardware)
- Putnam CD, Hammel M, Hura GL, Tainer JA (2007) "Solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution” Q. Rev. Biophys. 40, 191-285. (General SAXS)
Acknowledge SIBYLS
Please also acknowledge SIBYLS when using data generated at beamline 12.3.1.
These citations and acknowledgments are absolutely essential in keeping the MailinSAXS program funded and the beamline continually repaired and upgraded, as well as to provide access to experienced staff and the development of new technologies, such as SEC-SAXS.
Resources where to get additional information
Online Webinars
We regularly present online webinars to highlight new SAXS features, and how to address and solve specific problems that occur during SAXS data collection and processing:
Webinars